Cell-type diversity and regionalized gene expression in the planarian intestine
David J. Forsthoefel1,2,5, Nicholas I. Cejda1, Umair W. Khan2,3, Phillip A. Newmark2,4,5
Forsthoefel et al. eLife 2020;9:e52613.
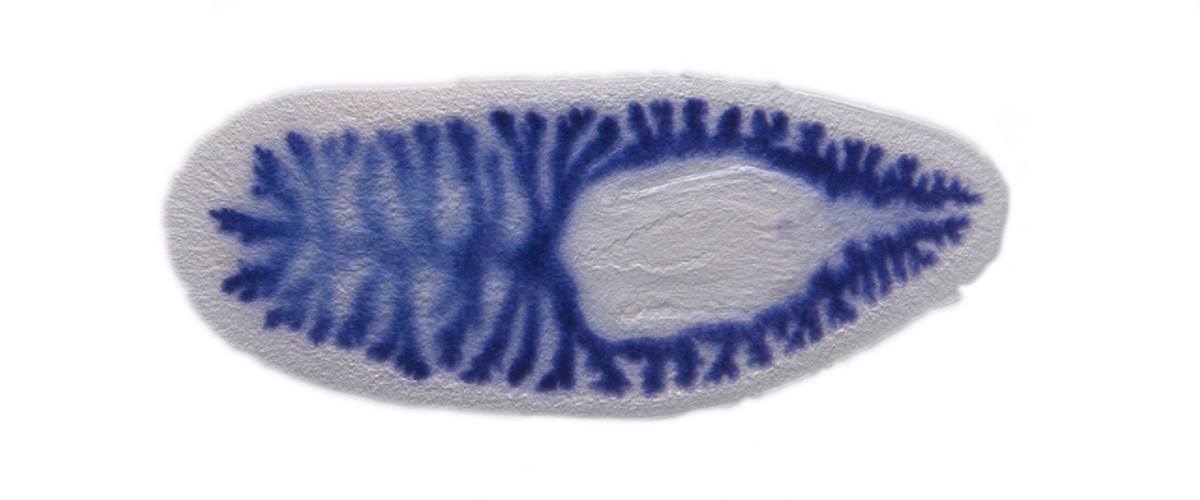
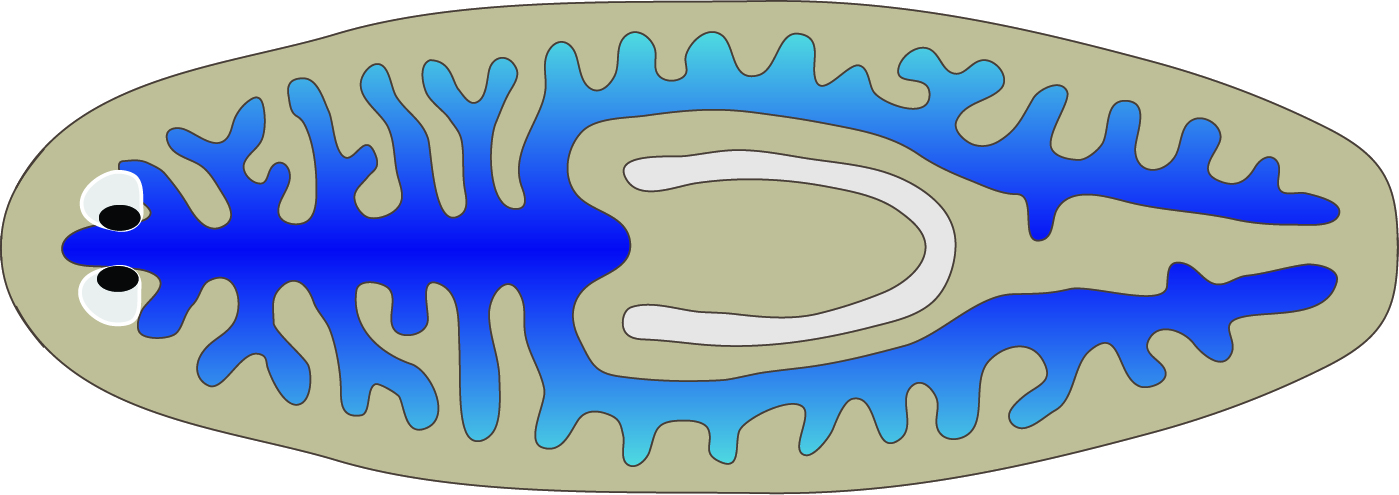
Proper function and repair of the digestive system are vital to most animals. Deciphering the mechanisms involved in these processes requires an atlas of gene expression and cell types. Here, we applied laser capture microdissection (LCM) and RNA-seq to characterize the intestinal transcriptome of Schmidtea mediterranea, a planarian flatworm that can regenerate all organs, including the gut. We identified hundreds of genes with intestinal expression undetected by previous approaches. Systematic analyses revealed extensive conservation of digestive physiology and cell types with other animals, including humans. Furthermore, spatial LCM enabled us to uncover previously unappreciated regionalization of gene expression in the planarian intestine along the medio-lateral axis, especially among intestinal goblet cells. Finally, we identified two intestine-enriched transcription factors that specifically regulate regeneration (hedgehog signaling effector gli-1) or maintenance (RREB2) of goblet cells. Altogether, this work provides resources for further investigation of mechanisms involved in gastrointestinal function, repair and regeneration.